Scientists have described mating-type determination mechanisms in Paramecium
Conjugation (or mating) of ciliates is a unique phenomenon among living beings. They have sex not for reproduction or pleasure — they seek to increase genetic variation. Scientists from St Petersburg University, together with colleagues from Poland and France, have studied the mating process in five sibling species of the Paramecium aurelia complex.
Their findings enabled them to describe genetic mechanisms behind this phenomenon. The research results have been published in the prestigious scientific journal Genome Biology and Evolution.
The mating process is one of the most important mechanisms for maintaining genetic variation in natural populations. The emergence of sexual reproduction turned out to be the most important evolutionary innovation that facilitated the evolution of eukaryotes. Paramecium is a well-known genus of ciliated protists with a complex system of ‘sexes’, or mating types. Paramecium reproduces asexually, by binary fission, which is not related to the mating process. During conjugation, Paramecium of compatible mating types exchange haploid nuclei, equivalent to gametes. The nuclei of each organism then fuse to form a diploid genome. This genome is stored in germline micronuclei of the exconjugants. It then undergoes large-scale rearrangements in the somatic macronucleus, including the elimination of virtually all non-coding DNA. Thus, there are two Paramecium cells before the formation of a conjugating pair, as well as after conjugation: no offspring are produced. However, by the time the two cells go their separate ways, both of them will have changed. Firstly, now they will have become genetically identical, and, secondly, they will both have acquired a new genetic identity, different from their ‘parental’ genotype.
I will draw an analogy with humans, whose sexual activity is not always associated with procreation as well. It is as if after sexual intercourse the two partners became identical twins. Moreover, the transformation is so radical that they change directly into their own offspring, as if being reincarnated in the same body but genetically different.
One of the authors of the study, professor in the Department of Microbiology of St Petersburg University, Dr. Alexey Potekhin
For several years, the scientists have studied mating-type systems in five closely related species of the ciliate Paramecium. They sought to find out how mating-type polymorphism is maintained in organisms that do not exhibit any sexual characteristics. ‘Ciliates are unorthodox about sex. For example, different Paramecium species have multiple, as well as binary mating types systems. The multitude of mating types significantly expands the choice of mating partners. Most importantly, mating occurs between cells of different genotypes, i.e. of different mating types. As for the binary systems, they are what we humans call «bisexual». As practice has shown, two sexes are quite enough,’ says Alexey Potekhin.
Some species of Paramecium have two mating types, O (Odd) and E (Even), which might be broadly called representatives of different ‘sexes’, since conjugation occurs only between cells of different mating types. However, the mechanisms of sex, or mating-type, determination remained unknown for a long time. Previous work in Paramecium tetraurelia revealed the key role of the mtA gene. It encodes a transmembrane protein that is specifically expressed in sexually reactive cells of type E. The mtA protein is directly involved in the species-specific recognition of type O reactive cells. For E expression, the mtA protein requires two other proteins, mtB and mtC, which encode factors that are necessary for mtA transcription in E reactive cells. All three proteins are specifically required for type E expression, whereas no gene is specifically required for type O expression.
To reconstitute functional genes in the somatic genome, ciliates rely on the programmed genome rearrangements, which include the excision of the unnecessary elements from coding sequences in the macronucleus. This excision event depends on a special class of small RNAs that scan the parental macronuclear genome to identify missing sequences and correlate it with the new one, which is to be formed after conjugation. Consequently, all DNA sequences that are absent from the parental macronucleus are eliminated. The parental macronucleus is then destroyed, and the new functional macronucleus is formed. ‘Meanwhile, the new macronuclear genotypes differ from the parental one, as it is expected to be as a result conjugation,’ notes Alexey Potekhin.
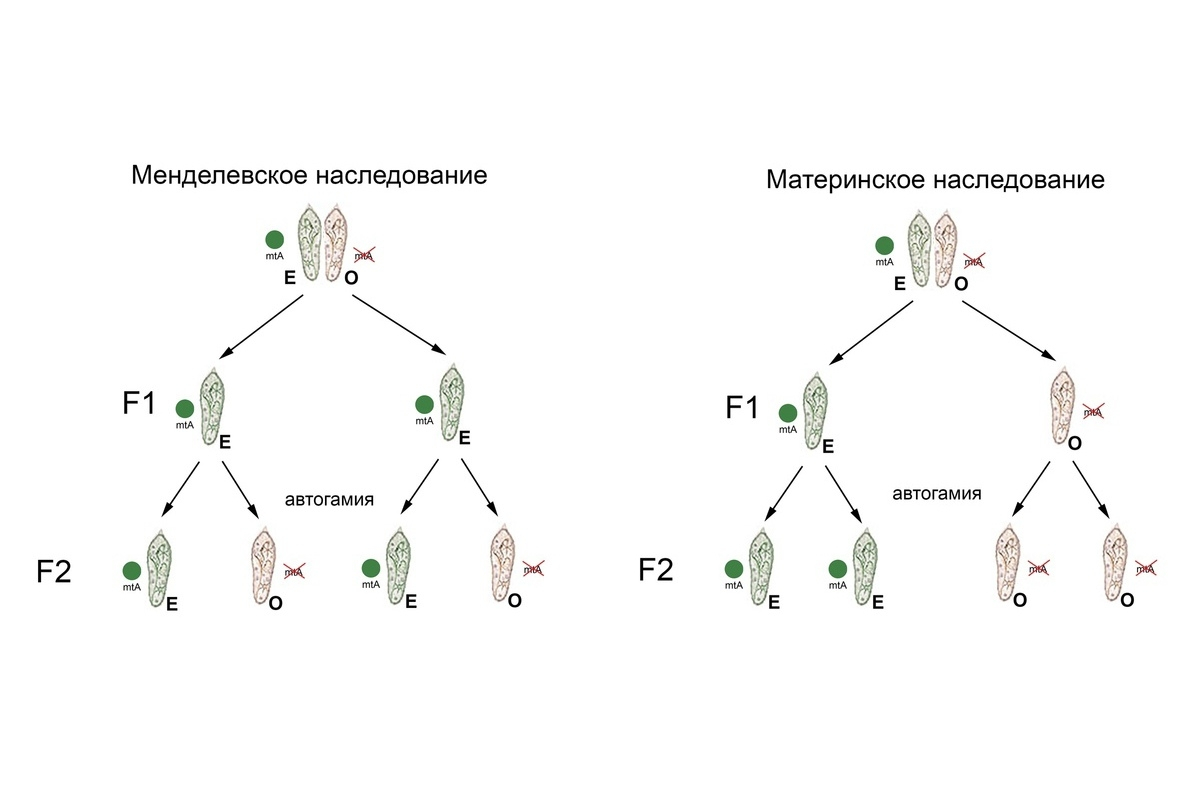
The mating type of Paramecium tetraurelia is inherited maternally by programmed genome rearrangements. In other words, each progeny acquires the mating type of its cytoplasmic parent. ‘A type O cell has an mtA gene that is inactivated during development, because the DNA segment, containing the mtA promoter (its transcription start site) and elements directly downstream, is reminiscent of transposons. The latter are also known as transposable elements, or jumping genes. They make up a large fraction of the non-coding DNA and can be eliminated from the macronuclear genome as unnecessary. In such case, this fragment will always be deleted in the next generations during macronuclear development. After all, it has already been lost from the «template». Thus, the mtA gene is inactivated, and the progeny inherits the mating type O,’ explains Alexey Potekhin.
The research was supported by the Russian Foundation for Basic Research (project No 19-04-00710a).
Among species with cytoplasmic inheritance of mating types, there are several species of the Paramecium aurelia complex: P. biaurelia, P. sexaurelia, P. septaurelia, P. octaurelia, P. decaurelia, and P. dodecaurelia. In the new study, the scientists have shown that these sibling species use the same mechanism, but different modes of mating-type determination. Several species use the same regulation mechanism as P. tetraurelia — an epigenetically inherited deletion of the fragment containing the mtA promoter and the elements directly downstream from the start site. Conversely, in P. sexaurelia, it is the last of the base pairs of the coding sequence of the mtA gene that undergo correction during genome rearrangements. In two other Paramecium species, P. biaurelia and P. septaurelia, the mtB gene was found to be rearranged differently — some fragments of the mtB gene may get deleted leading to the mtB deficiency. Due to the absence of functional mtB, the mtA gene cannot function properly. Thus the absence of functional mtB is what determines mating type O in these species.
When I was a student 25 years ago, I was amazed that within a small group of ciliates there are three different modes of mating-type determination and inheritance. Such evolutionary plasticity of mating-type systems is unique and cannot be found in multicellular organisms. At the time, it was an absolute mystery, because then no one had an idea of genome scanning, nor of mating-type genes.
Professor in the Department of Microbiology of St Petersburg University, Dr. Alexey Potekhin
‘Our research team has been able to make a great contribution to the study of this phenomenon, and I am pleased with the scientific results we have achieved. It has been a real scientific quest,’ stresses Alexey Potekhin.
Mechanisms of mating-type determination in different species of Paramecium aurelia complex. The mtA gene is specifically required for type E expression, while its transcription is provided by the mtB and mtC factors, or unknown proteins with the same function. It is shown that deletions in P. biaurelia and P. septaurelia type O clones remove different parts of the coding sequence of the mtB gene.
In Paramecium tredecaurelia, mating types are not inherited cytoplasmically, but follow a classical pattern of Mendelian inheritance. It has been shown that in that species, O and E mating types are genetically determined by the loss of a single nucleotide in the mtA promoter in type E clones. The genome of P. tredecaurelia contains no functional mtB gene; therefore, another transcription factor plays its role. The conducted experiments enabled the scientists to establish that the transcription factor binding site is located in the promoter region with a discovered one nucleotide deletion, while the mtA gene is activated by such a promoter. If there is no deletion, then the site is not recognised and the mtA gene is not expressed during sexual reactivity. The same binding site in the mtA promoter is also observed in three more species of the Paramecium aurelia complex, with all of them lacking the mtB gene.
‘This implies that in the evolution of these sibling species, there occurred several evolutionary transitions between mating-type determination systems. Despite different molecular solutions, the result was always the same: in a natural population, there will be cells of both mating types. The evolutionary history of the Paramecium aurelia illustrates that any regulation mechanism for maintaining mating-type polymorphism in cell populations can take hold, involving mating-type biased gene expression and programmed genome rearrangements. This shows how important the mating process is even for unicellular organisms,’ notes Alexey Potekhin.