International conference BiATA celebrates its jubilee
St Petersburg University has held the fifth international conference ‘Bioinformatics: from Algorithms to Applications’ (BiATA 2021). It is held annually by the Center for Algorithmic Biotechnology at St Petersburg University.
‘In 2021, Russian scientists presented a wide range of rigorous reports that aroused curiosity and interest in scientific community. It is good evidence that bioinformatics is becoming an indispensable part of science in Russia and our scientists are becoming more and more competitive. In previous years, most of those who participated in the conference were primarily scientists from abroad. Additionally, the conference has significantly broadened its geography across Russia. We have participants from St Petersburg, Moscow, Novosibirsk, Ufa, and Tver to name but a few. The conference has survived the harsh
COVID-19 reality. It is important for us as BiATA is the only conference of its kind in St Petersburg and one of few across Russia’, said Alla Lapidus, Chairperson of the BiATA 2021 Programme Committee.
The conference featured a report ‘NGS data for natural GMOs discovery and investigation’ by Tatiana Matveeva, Doctor of Biology and Professor at St Petersburg University.
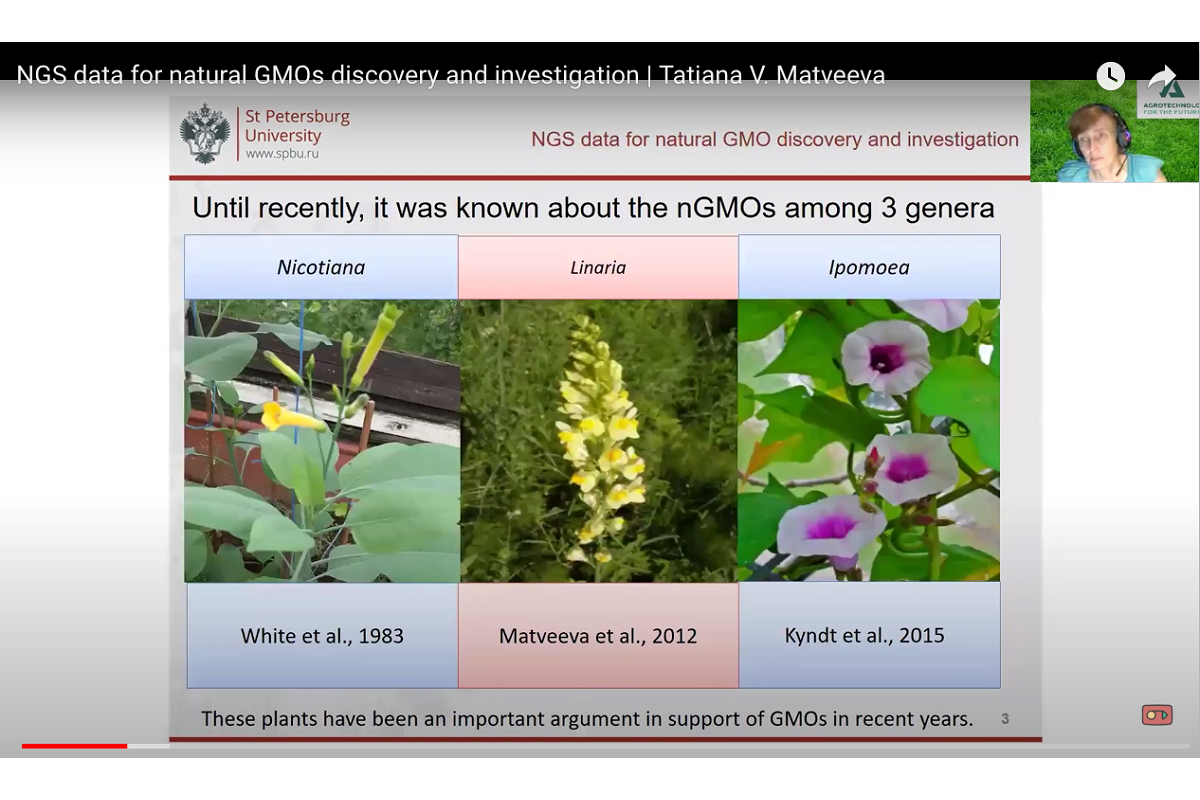
Naturally transgenic plants are species which were subjected to Agrobacteriummediated transformation millions years ago. Agrobacterium-mediated transformation is associated with horizontal gene transfer, i.e. insertion of T-DNA of the Ti plasmid in the plant genome.
She spoke about the known species that had DNA sequences in their genomes, homologous to agrobacterial ones, and continued to pass them on from generation to generation. For example, it can be found in Nicotiana, Ipomea, Linaria, and over 30 species of wild and cultivated plants.
Today, agrobacterium-mediated transformation is the most common method for obtaining commercial lines of transgenic plants. Despite the fact that this method is based on a natural vector system, genetically modified organisms (GMOs) are constantly attacked as organisms alien to nature.
Naturally transgenic plants are key to studying the consequences of distribution of GMOs, said Professor Matveeva. They can enable us to gain a deeper insight into what will happen to GMOs after a long period of time and what are the functions of those parts of the T-DNA that can be found in the genomes of some plants.
Varvara Tvorogova, Senior Research Associate in the Department of Genetics and Biotechnology at St Petersburg University, delivered a report ‘The search of somatic embryogenesis regulators in plants'. The research was guided by Liudmila Lutova, Professor at St Petersburg University.
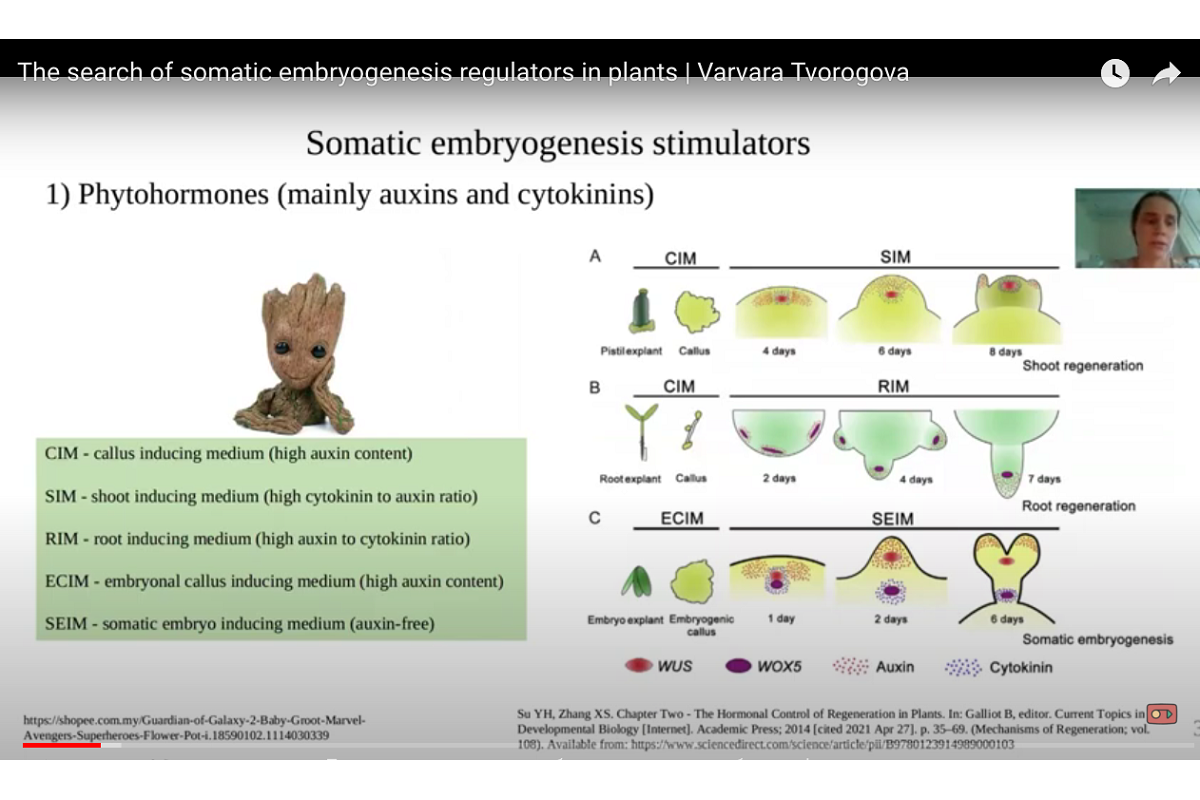
Somatic embryogenesis (SE) is the way of regeneration of plant, during which the vegetative cells form embryos, which potentially can develop into the new plant. This process is used in the plant biotechnology for genome modification and plant breeding. The ability of plant to form somatic embryos in vitro depends on cultivation conditions and also on the plant genotype. The investigation of genes regulating this process can therefore help to improve the SE protocols.
‘We search for SE regulators among the genes from WUSCHEL-RELATED HOMEOBOX (WOX) and NUCLEAR FACTOR Y (NF-Y) families, encoding transcription factors, as well as among the genes from CLAVATA3/EMBRYO SURROUNDING REGION (CLE) family, encoding peptide hormones, using Medicago truncatula (barrel medic) as model object’, said Varvara Tvorogova.
Scientists found that MtWOX9-1 and STENOFOLIA genes from WOX family are able to stimulate SE in M. truncatula when overexpressed. Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA. Gene overexpression is the process which leads to the abundant target gene expression in transgenic or recombinant organisms. Edited plants with loss of MtWOX9-1 function are sterile, which supports the role of this gene in the embryogenesis.
We also found one possible suppressor of SE among CLE genes, which can be used as potential target for editing to obtain the plants with high capacity for regeneration.
Varvara Tvorogova, Senior Research Associate in the Department of Genetics and Biotechnology at St Petersburg University
The research groups headed by Tatiana Matveeva and Varvara Tvorogova are part of the world-class research centre 'Agrotechnologies of the Future'. The centre was established as part of the national project 'Science' by: St Petersburg University; Russian State Agrarian University – Moscow Timiryazev Agricultural Academy; Federal Research Centre
'Fundamentals of Biotechnology' of the Russian Academy of Sciences; Federal Research Centre ‘Informatics and Management’ of the Russian Academy of Sciences; All-Russian Research Institute of Agricultural Microbiology; Dokuchaev Soil Institute; and Federal Research Centre N. I. Vavilov All-Russian Institute of Plant Genetic Resources (VIR).
Held online in 2021, BiATA brought together scientists from France, Germany, Portugal, Chile, the USA, South Korea, the Czech Republic, India, England, Egypt, Belarus, Thailand, and the Philippines.
The University scientists analysed how genomes work in various stages of life cycle of the parasitic flatworms, Trematoda, and female body parts of the parasitic Rhizocephala. This was the focus of the report by Maksim Nesterenko, Junior Research Associate in the Department of Zoology of Invertebrates at St Petersburg University. Among what makes Trematoda different is their complex life cycle. It is associated with the rotation of the sex generations. Each generation is characterised by specific ontogenesis, i.e. development of an individual organism.
Unlike Trematoda, the life cycle of Rhizocephala is rather simple. It is associated with one generation only. Yet the female body has significantly changed throughout the evolution. Now it resembles a root system of a plant with specific parts. To gain a better understanding of molecular differences in the stages of the cycles of Trematoda and body parts of Rhizocephala, scientists used the following techniques: philostratigraphy and evolutionary transcriptomes. These methods describe the work of the groups of genes that originate at different stages of evolution of the organisms. They revealed the statistically significant differences in the samples in terms of how active the ‘old’ and ‘new’ genes were. 'Yet we should take precautions in interpreting the age indexes of the transcriptomes’, said Maksim Nesterenko. He added that the data was partially analysed at the resource centre ‘Computing Centre’ at St Petersburg University. The obtained results will be further analysed in his dissertation to gain a degree of candidate of sciences.
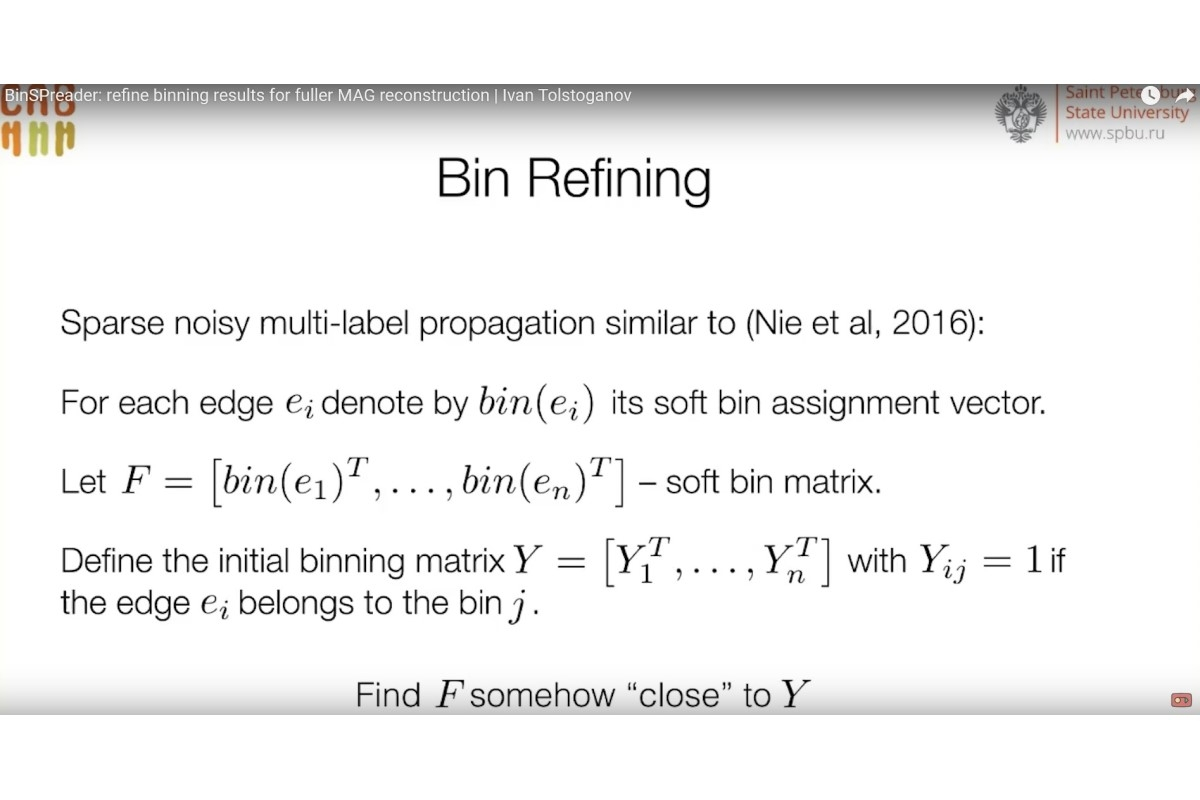
Ivan Tolstoganov is Junior Research Fellow at the Center for Algorithmic Biotechnology at St Petersburg University. He spoke about a new tool BinSPreader. . He said that analysing metagenome, i.e. genetic content of a group of microorganisms, is essential to study a community of uncultivated microorganisms. An important part of the analysis of metagenomic sequencing is binning, i.e. process of grouping reads or contigs and assigning them to individual genome.
'Today's tools for binning are far from being able to classify short nucleotide sequences that contain interesting data, e.g. antibiotic resistance genes or differences in strains’, said Ivan Tolstoganov.
The tool developed by us to enhance the quality of binning can significantly add to the classification of short sequences and partly correct errors in classification of long sequences of the initial binning.
Ivan Tolstoganov, Junior Research Fellow at the Center for Algorithmic Biotechnology at St Petersburg University
His report was followed by a number of enquiries to obtain a beta-version of BinSPreader at the Center for Algorithmic Biotechnology at St Petersburg University.
The Center for Algorithmic Biotechnology at St Petersburg University is actively collaborating with the European Bioinformatics Institute (EMBL-EBI), said Alla Lapidus. EMBL-EBI is among the world's flagship laboratory for the life sciences. 'The Center for Algorithmic Biotechnology at St Petersburg University is a world-recognised centre’, said Alla Lapidus, Chairperson of the BiATA 2021 Programme Committee.
The conference featured a workshop ‘Analysing metagenomic assemblies using MGnify’. Scientists could learn how to use MGnify. i.e. one of the most innovative platforms for analysis of metagenomics assemblies that was developed by the European Bioinformatics Institute (EMBL-EBI) in collaboration with the Center for Algorithmic Biotechnology at St Petersburg University.